Example 3 - High-level, guided processing (using gesture data)
This example shows the usage of class DataProcessingManager, a high-level, guided processing interface with accepted EMG processing conventions. This is the recommended way of EMG processing using this package.
The data used in this example contains two files of subject #10 in “EMG data for gestures” from the UC Irvine Machine Learning Repository. In particular, the data was collected in eight channels from the forearm of the subject while performing series of static hand gestures.
Package preparation
We install and import needed packages.
[1]:
!pip install pyemgpipeline -q
[2]:
import os
import numpy as np
from matplotlib.figure import SubplotParams
import pyemgpipeline as pep
Data preparation
First download the raw data from the UC Irvine Machine Learning Repository.
When finished, the compressed file “EMG_data_for_gestures-master.zip” (~17MB) is saved in the “uci_gestures” subfolder.
[3]:
!rm -rf uci_gestures
!wget -q https://archive.ics.uci.edu/ml/machine-learning-databases/00481/EMG_data_for_gestures-master.zip -P ./uci_gestures
Uncompress the downloaded file. This example will only use two files of subject 10, which are in the archived folder “EMG_data_for_gestures-master/10”.
[4]:
# !unzip -q uci_gestures/EMG_data_for_gestures-master.zip -d ./uci_gestures #This command extracts all files.
!unzip -q uci_gestures/EMG_data_for_gestures-master.zip EMG_data_for_gestures-master/10/* -d ./uci_gestures #This command extracts only the file needed in this example.
Signal data of each trial should be stored in a 2d ndarray with shape (n_samples, n_channels), where each column represents data of one channel. Data of multiple trials are organized in a list.
First we set up basic information of the data, including data path, channel names, sample rate, etc.
[5]:
data_folder = 'uci_gestures/EMG_data_for_gestures-master/10'
data_filenames = ['1_raw_data_11-08_21.03.16.txt', '2_raw_data_11-10_21.03.16.txt']
trial_names = ['trial 1', 'trial 2']
channel_names = ['channel1', 'channel2', 'channel3', 'channel4', 'channel5', 'channel6', 'channel7', 'channel8']
sample_rate = 1000
Signal data of multiple trials are stored in all_data of type list, where each element of the list is a 2d ndarray. Since the raw data provides the timestamp information, it is also extracted in all_timestamp of type list, where each element is a 1d ndarray.
[6]:
all_timestamp = []
all_data = []
for fn in data_filenames:
data = np.genfromtxt(os.path.join(data_folder, fn), delimiter='\t', skip_header=1)
all_timestamp.append(data[:, 0] / sample_rate)
all_data.append(data[:, 1:9])
[7]:
all_timestamp
[7]:
[array([1.0000e-03, 5.0000e-03, 6.0000e-03, ..., 6.4554e+01, 6.4555e+01,
6.4557e+01]),
array([1.0000e-03, 5.0000e-03, 6.0000e-03, ..., 6.3954e+01, 6.3955e+01,
6.3956e+01])]
[8]:
all_data
[8]:
[array([[-2.e-05, -1.e-05, -3.e-05, ..., -1.e-05, 1.e-05, 1.e-05],
[-2.e-05, -1.e-05, -3.e-05, ..., -1.e-05, 1.e-05, 1.e-05],
[-2.e-05, -1.e-05, -3.e-05, ..., -1.e-05, 1.e-05, 1.e-05],
...,
[ 0.e+00, 3.e-05, 4.e-05, ..., -3.e-05, 0.e+00, 0.e+00],
[ 0.e+00, 3.e-05, 4.e-05, ..., -3.e-05, 0.e+00, 0.e+00],
[ 0.e+00, 3.e-05, 4.e-05, ..., -3.e-05, 0.e+00, 0.e+00]]),
array([[-2.e-05, -5.e-05, -4.e-05, ..., -2.e-05, 1.e-05, 0.e+00],
[-2.e-05, -5.e-05, -4.e-05, ..., -2.e-05, 1.e-05, 0.e+00],
[-2.e-05, -5.e-05, -4.e-05, ..., -2.e-05, 1.e-05, 0.e+00],
...,
[ 1.e-05, 1.e-05, 0.e+00, ..., -1.e-05, -1.e-05, -1.e-05],
[ 1.e-05, 2.e-05, 5.e-05, ..., -2.e-05, -1.e-05, -1.e-05],
[ 1.e-05, 2.e-05, 5.e-05, ..., -2.e-05, -1.e-05, -1.e-05]])]
[9]:
print('shapes of all_timestamp:', all_timestamp[0].shape, all_timestamp[1].shape)
print('shapes of all_data:', all_data[0].shape, all_data[1].shape)
shapes of all_timestamp: (61641,) (61448,)
shapes of all_data: (61641, 8) (61448, 8)
We also set up the parameters for plotting with an instance of class EMGPlotParams. Note that the whole setting configures the plotting of a single trial. Plotting of all trials use the same setting.
[10]:
emg_plot_params = pep.plots.EMGPlotParams(
n_rows=2,
n_cols=4,
fig_kwargs={
'figsize': (16, 3),
'subplotpars': SubplotParams(top=0.8, wspace=0.1, hspace=0.4),
}
)
Data processing
First we create an instance of class DataProcessingManager.
[11]:
mgr = pep.wrappers.DataProcessingManager()
We use method set_data_and_params to set or reset signal data all_data, timestamp data all_timestamp, and other information, to the data processing manager instance.
[12]:
mgr.set_data_and_params(all_data, hz=sample_rate,
all_timestamp=all_timestamp, trial_names=trial_names,
channel_names=channel_names, emg_plot_params=emg_plot_params)
Note that the above method also initializes five processing steps as default in case they are not set yet. The steps can be examined as follows:
[13]:
mgr.show_current_processes_and_related_params()
---- Current processes and related parameters ----
DC offset remover : No parameters
Bandpass filter : hz = 1000, bf_order = 4, bf_cutoff_fq_lo = 10, bf_cutoff_fq_hi = 450
Full wave rectifier : No parameters
Linear envelope : hz = 1000, le_order = 4, le_cutoff_fq = 6
End frame cutter : n_end_frames = 30
The data processing manager instance contains instances of all processors (which can be None), including DCOffsetRemover, BandpassFilter, FullWaveRectifier, LinearEnvelope, EndFrameCutter, AmplitudeNormalizer, and Segmenter. All of them can be set or reset.
For example, we reset the BandpassFilter processor with different parameters and show current processes again.
[14]:
mgr.set_bandpass_filter(pep.processors.BandpassFilter(hz=sample_rate, bf_order=4, bf_cutoff_fq_lo=5, bf_cutoff_fq_hi=470))
mgr.show_current_processes_and_related_params()
---- Current processes and related parameters ----
DC offset remover : No parameters
Bandpass filter : hz = 1000, bf_order = 4, bf_cutoff_fq_lo = 5, bf_cutoff_fq_hi = 470
Full wave rectifier : No parameters
Linear envelope : hz = 1000, le_order = 4, le_cutoff_fq = 6
End frame cutter : n_end_frames = 30
We then set the AmplitudeNormalizer processor, which will calculate the maximum amplitude of each channel across trials for normalization.
[15]:
mgr.set_amplitude_normalizer(pep.processors.AmplitudeNormalizer())
mgr.show_current_processes_and_related_params()
---- Current processes and related parameters ----
DC offset remover : No parameters
Bandpass filter : hz = 1000, bf_order = 4, bf_cutoff_fq_lo = 5, bf_cutoff_fq_hi = 470
Full wave rectifier : No parameters
Linear envelope : hz = 1000, le_order = 4, le_cutoff_fq = 6
End frame cutter : n_end_frames = 30
Amplitude normalizer : No parameters
The process_all method is responsible for applying all processors in the predefined sequence. With the parameter is_plot_processing_chain being True, the original data and intermediate data after each step will be displayed.
[16]:
c = mgr.process_all(is_plot_processing_chain=True)
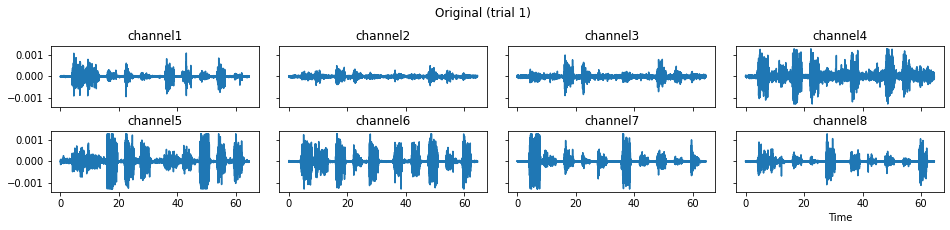
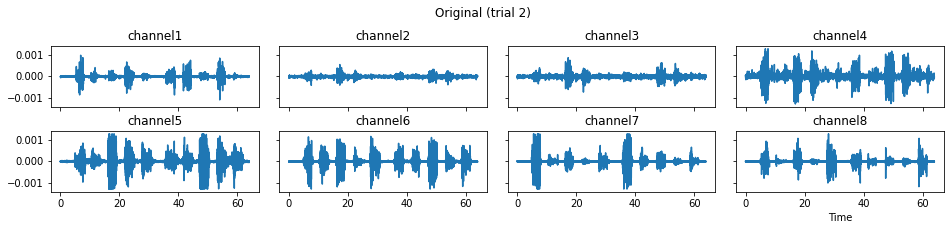
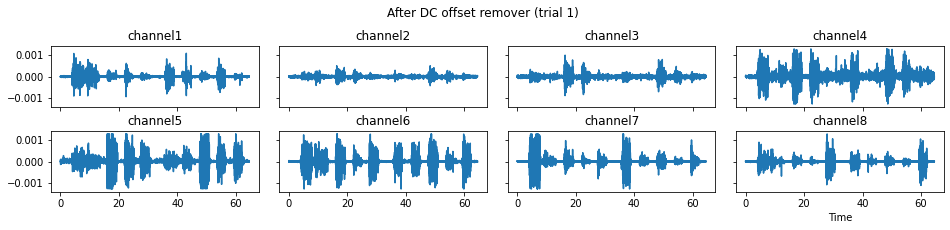
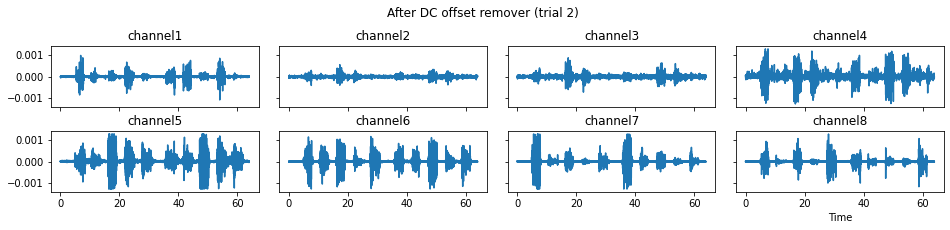
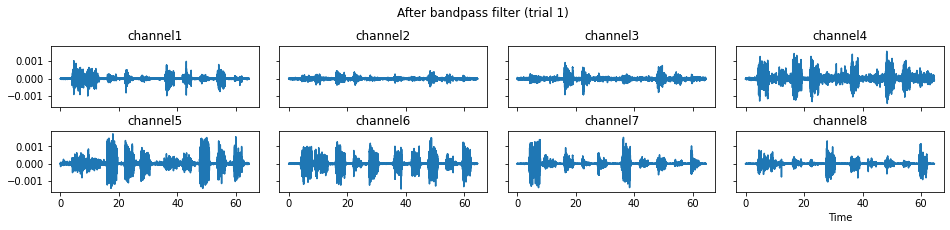
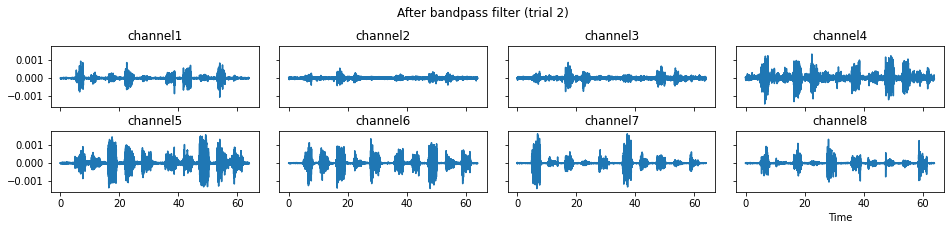
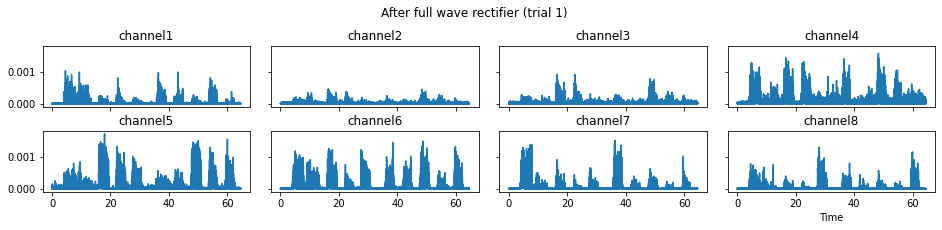
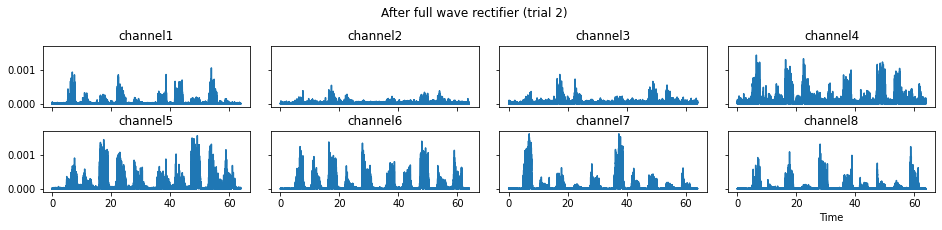
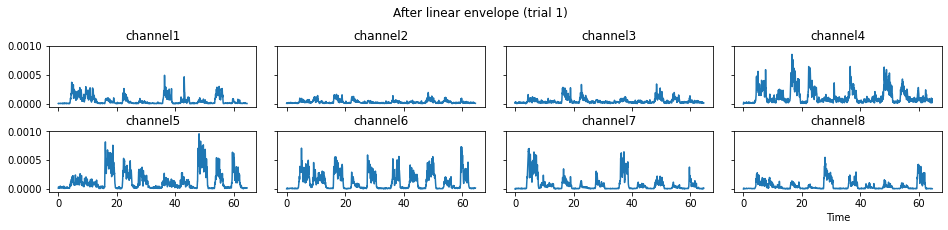
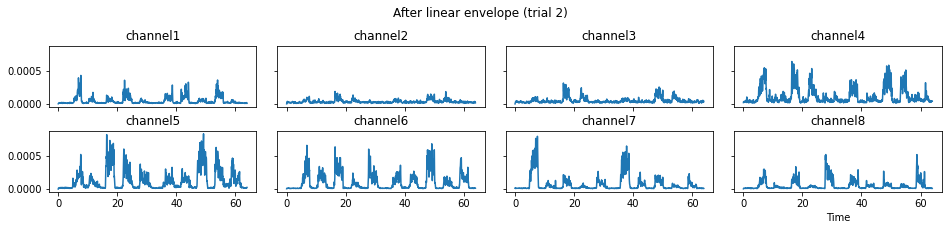
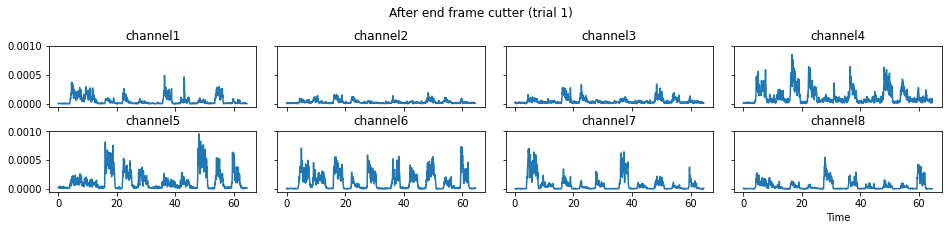
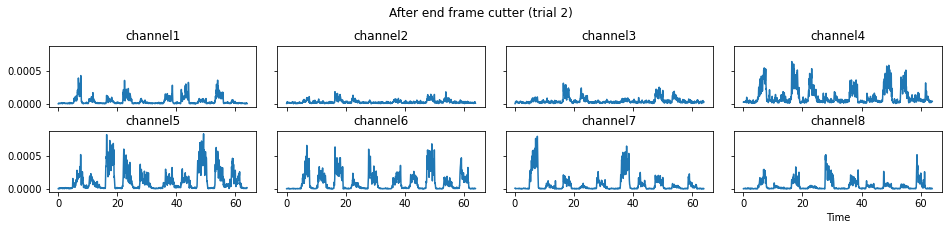
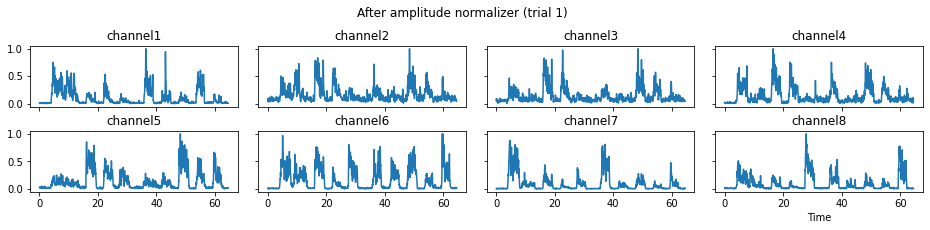
For demonstration purpose, assume the segment of interest of trial 2 is in the time range (15, 28). We can add the Segmenter processor and the time range to the data processing manager as the following.
[17]:
all_beg_ts = [0, 15]
all_end_ts = [999, 28]
mgr.set_segmenter(pep.processors.Segmenter(), all_beg_ts, all_end_ts)
mgr.show_current_processes_and_related_params()
---- Current processes and related parameters ----
DC offset remover : No parameters
Bandpass filter : hz = 1000, bf_order = 4, bf_cutoff_fq_lo = 5, bf_cutoff_fq_hi = 470
Full wave rectifier : No parameters
Linear envelope : hz = 1000, le_order = 4, le_cutoff_fq = 6
End frame cutter : n_end_frames = 30
Amplitude normalizer : No parameters
Segmenter : No parameters
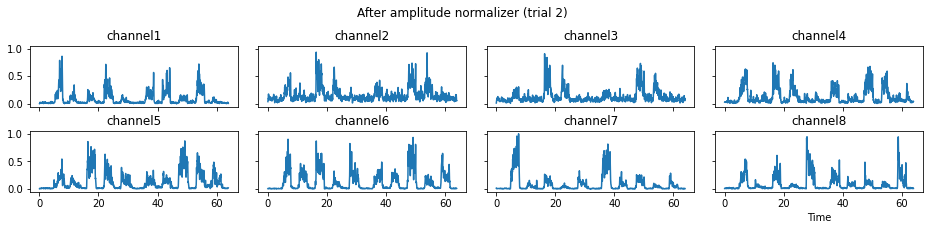
We now run the process_all method again with the updated processing steps. When is_plot_processing_chain is True, k_for_plot=1 indicates that only trial index 1 (i.e., trial 2) will be plotted. The segmentation result is shown in the last part of the figures.
[18]:
c = mgr.process_all(is_plot_processing_chain=True, k_for_plot=1)
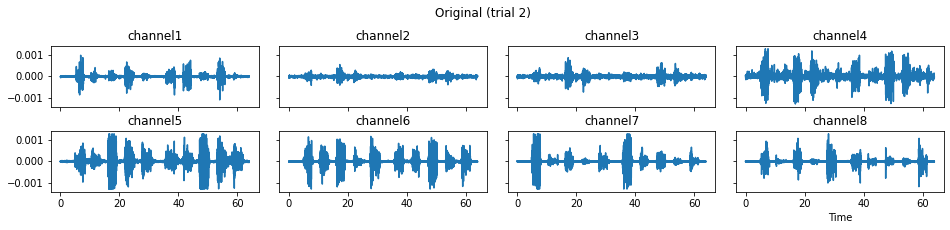
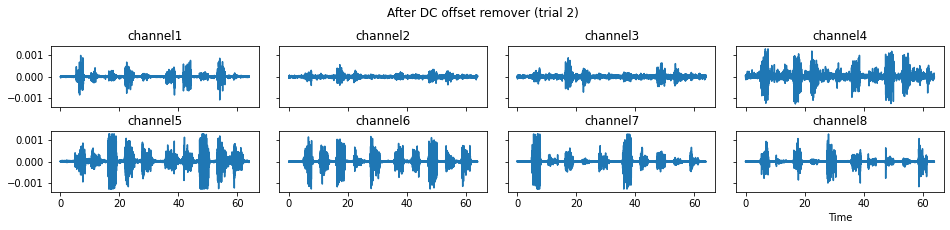
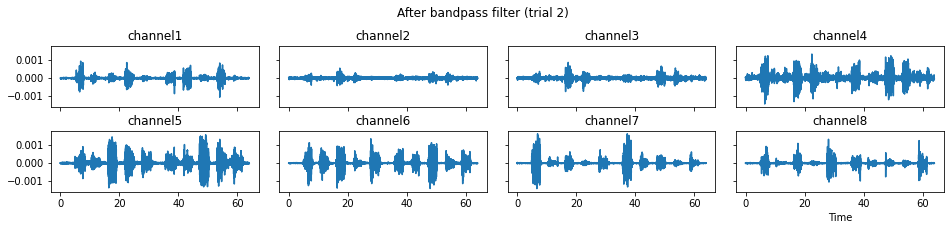
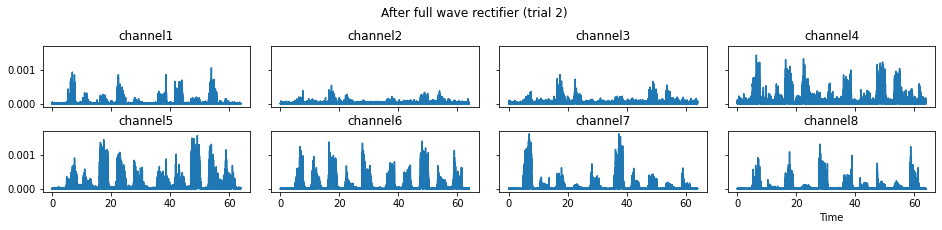
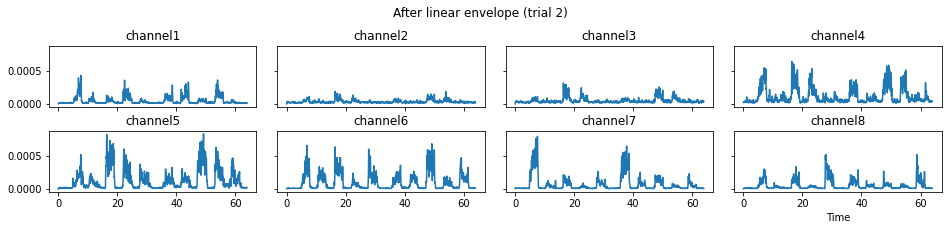
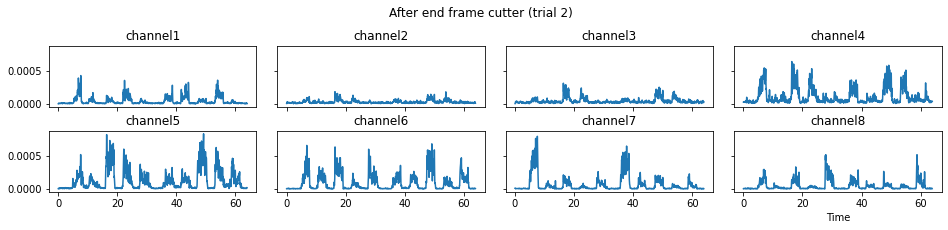
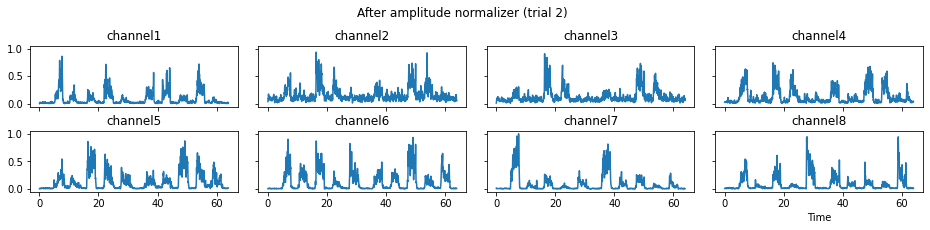
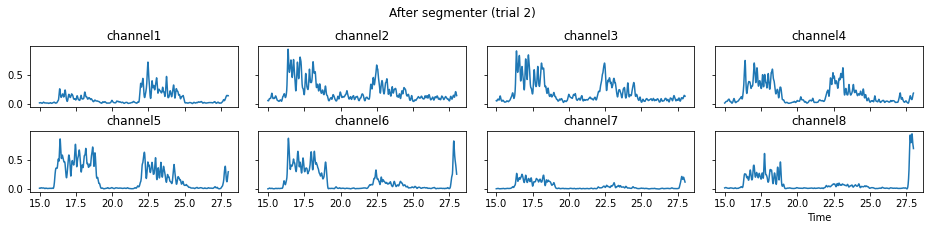
The processed results from method process_all is saved in c of type EMGMeasurementCollection, which possesses methods plot, export_csv, and the indexing operator [ ] for furtuer use.
[19]:
isinstance(c, pep.wrappers.EMGMeasurementCollection)
[19]:
True